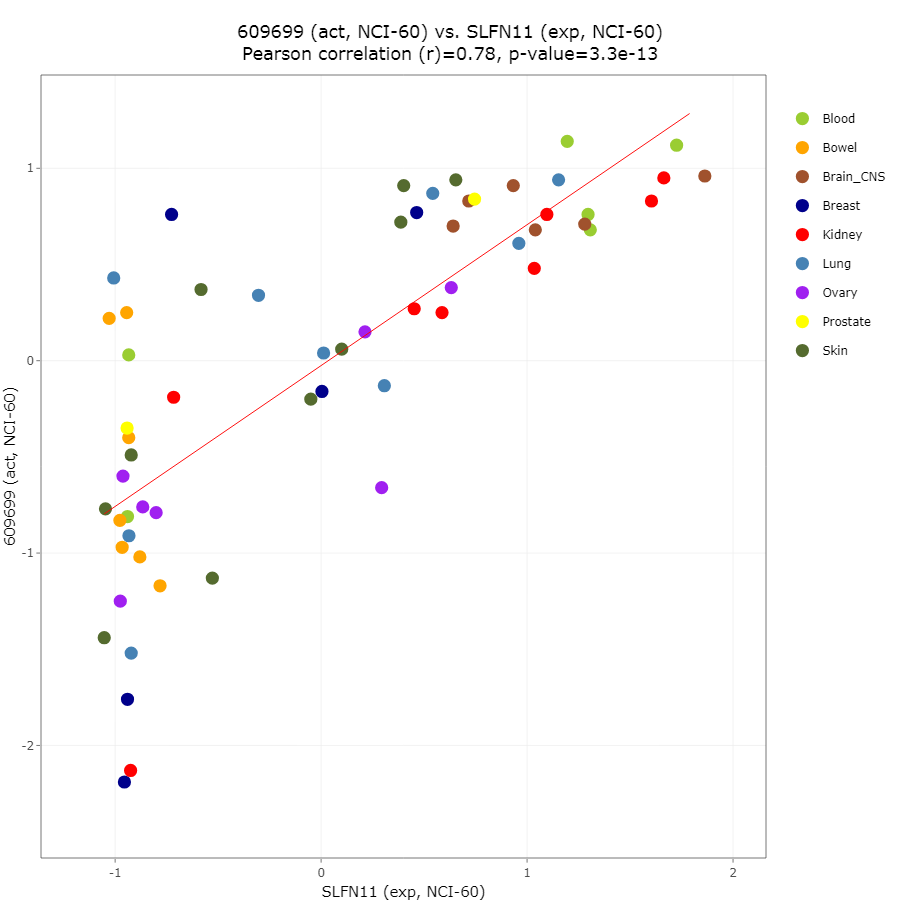
SLFN11 (Schlafen 11) belongs to a family of slfn that only exists in mammals. As schlafen means “sleep” in German, SLFN11 was “sleeping” until the two breakthrough papers came out in 2012. One paper from Dr. Yves Pommier lab (Zoppoli et al, PNAS. 2012) revealed that that mRNA expression level of SLFN11 is the most highly correlated to the sensitivity to a broad type of DNA-damaging drugs in large cancer cell line database of the NCI-60 (Figure 1). The DNA-damaging drugs connected to SLFN11 include topoisomerase I inhibitors (camptothecin, topotecan, irinotecan and indenoisoquinoline), topoisomerase II inhibitors (etoposide, mitoxantrone and doxorubicin), platinums (cisplatin, carboplatin and oxaliplatin), and DNA synthesis inhibitors (gencitabine, cytarabine and nucleoside analogs). The other group from Broad Institute led by Dr. Levi A. Garraway also reported the similar correlation between SLFN11 expression and the sensitivity to irinotecan using 947 cancer cell lines (Barretina et al., Nature. 2012). These findings are amazing because they imply that SLFN11 expression level alone could be a predictive marker for the drug sensitivity to various DNA damaging agents.
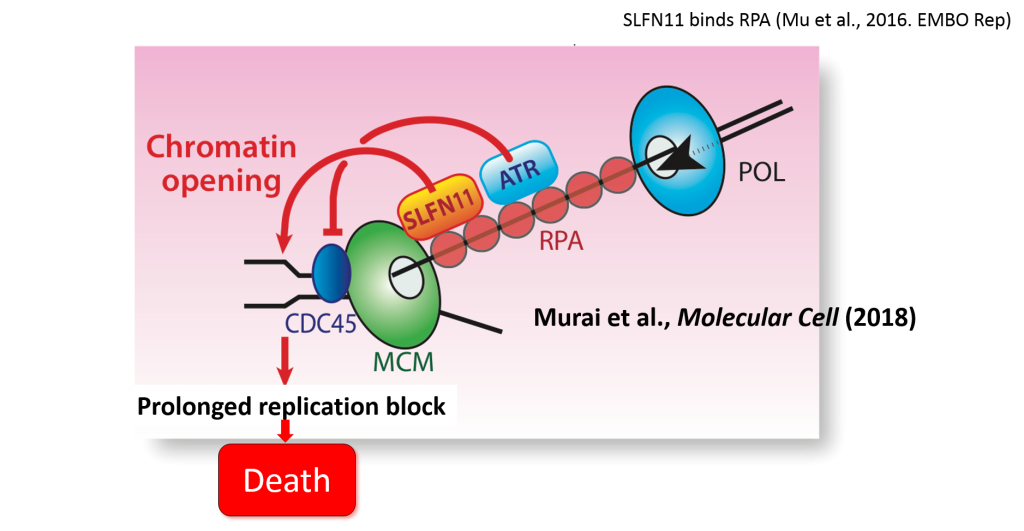
Hereafter, studies for SLFN11 have gradually expanded to the Clinic and the basic science. The mechanisms by which SLFN11 augments the toxicity of DNA-damaging drugs was recently discovered by Dr. Pommier group (Figure 2) (Murai et al., Mol Cell. 2018).
Clinical implication of SLFN11
Although DNA-damaging agents such as cisplatin, oxaliplatin and etoposide are used for decades as a first choice drug for some cancers, there is no clinical biomarker that can predict drug efficiency. DNA-damaging agents frequently confer severe side-effects such as myelosupression, infection, alopecia and diarrhea. Hence, it is critically important to select patients who can receive benefits (tumor regression, longer survival) from the hazardous drugs. Since SLFN11 augments the anti-tumor effect of DNA-damaging agents, SLFN11 can be a predictive biomarker for these drugs. Importantly, SLFN11 expression is very low in half of cancer cell lines accessed by lar1036ge cancer cell line database (Figure 3), it implies that SLFN11 can select ~50% of patients
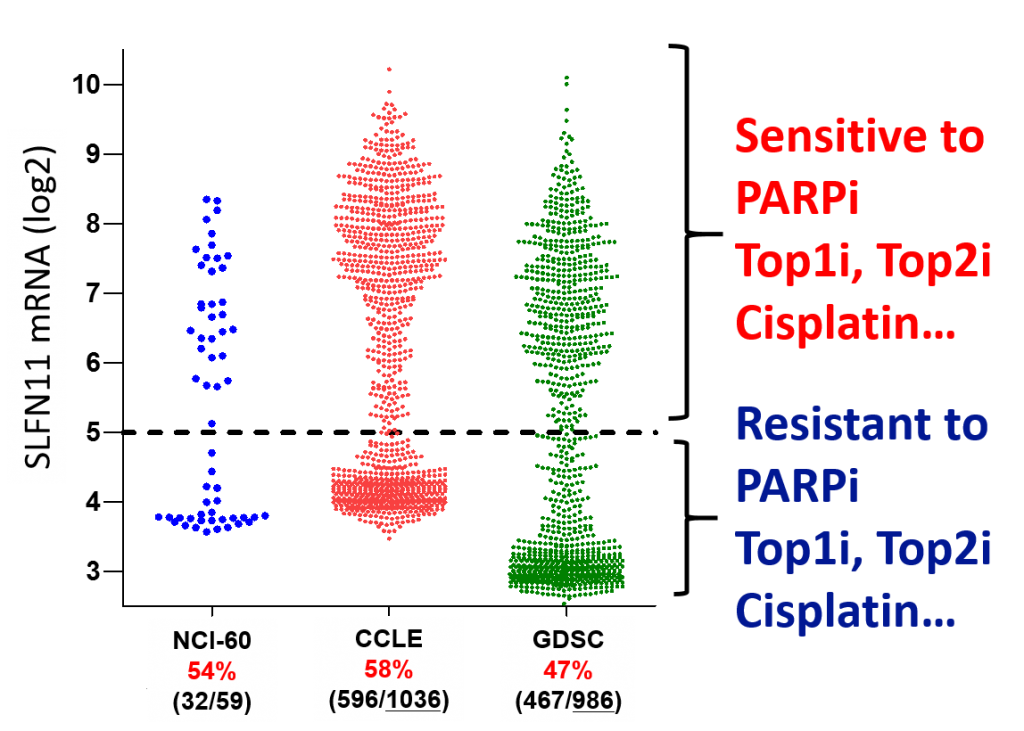
Figure 3. The clear ON and OFF distribution pattern of SLFN11 across three cancer cell line database. The numbers of SLFN11 “ON” cell lines/ total cell lines are shown within parenthesis. Percentage of SLFN11 “ON” cells are shown by red. Data are obtained from CellMinerCDB, and modified.